
experimental evolution with yeast
Saccharomyces cerevisiae (brewer's yeast) is a species with an impressive amount of genetic diversity. We use S. cerevisiae strains collected from locations around the world in laboratory selection experiments to simulate real-world scenarios.
These experiments allow us to test hypotheses about:
-
how populations will adapt to climate change
-
physiological consequences of adaptation to stress
-
the origins and fates of adaptive alleles
-
the repeatability of evolution
-
advantages of recombination and outcrossing
-
the evolution of aging and reproductive longevity

photo: molly burke
This photo shows yeast cells that have been stained so that you can see their bud scars - these are the visible scars of cell division. Cells with more bud scars are older than cells with fewer bud scars.
experimental evolution with Drosophila
While we work with yeast, we collaborate with like-minded labs doing selection experiments with other organisms. We are involved in ongoing projects in which populations of the fruit fly Drosophila melanogaster are selected for physiological and fitness traits.
We use phenotype and genome data from these populations to investigate evolution in real time, and to assess the limits of what we can learn from these experiments.

Here we see Molly thinking hard about flies (photo cred: Steve Zylius)
genetics and evolution of aging
Aging is a complex disease determined by both environmental and genetic factors, and their interactions. Genome-wide association studies (GWAS) allow the identification of genetic variants relevant for aging and age-related disease. But, there are many challenges to studying human aging with this method, mainly because it is difficult to assemble large panels of very old individuals, with appropriate controls.
GWAS in laboratory experiments with model organisms can circumvent some of these challenges. Our goal is to better understand the genetics underlying aging and longevity in multiple species. Ongoing projects involve both flies and yeast. By analyzing the DNA and RNA of the individuals that survive the longest in a given cohort, we aim to identify genetic variants that play key roles in determining longevity.
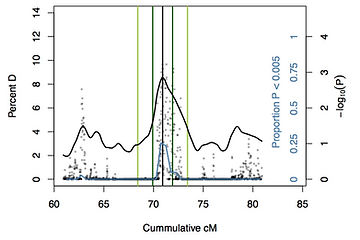
This figure shows a region of candidate SNPs on chromosome 2L associated with extreme longevity in Drosophila melangaster.
(published 2014 in Genome Biology and Evolution)